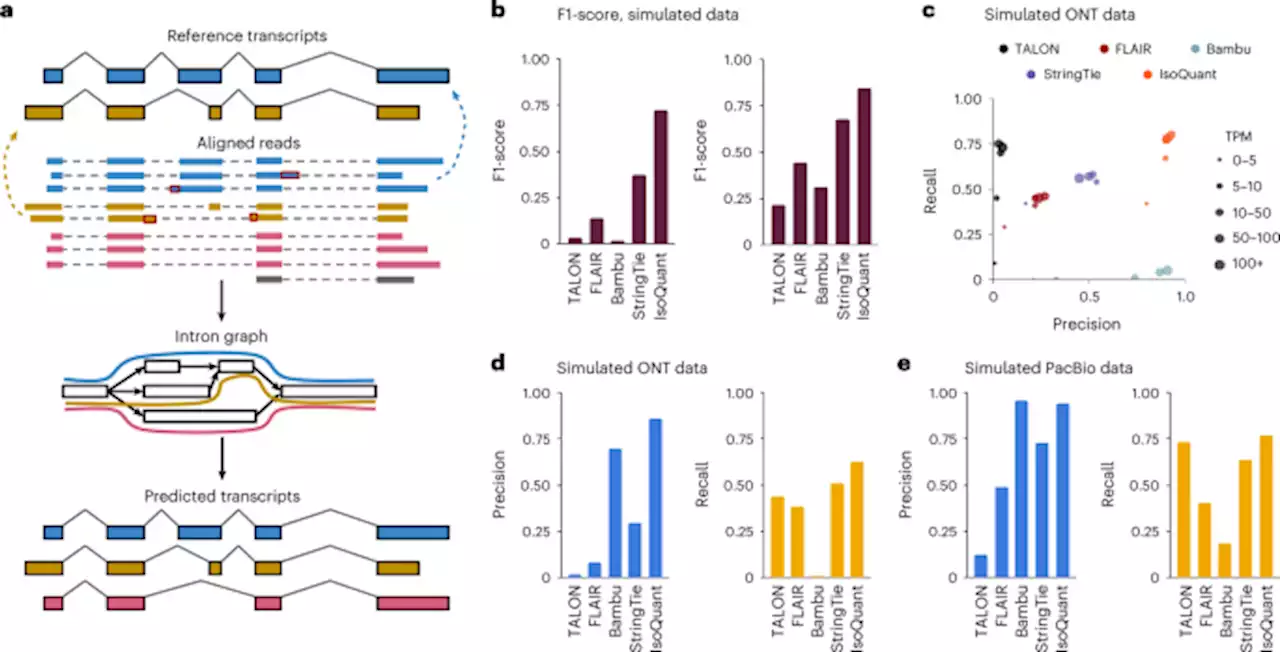
Accurate isoform discovery with IsoQuant using long reads
To mimic real-life situations and assess the ability of an algorithm to predict novel transcripts, we created reduced gene annotations by removing a fraction of expressed isoforms. First, we define a subset of true expressed transcripts that contributed to at least one read during the simulation. Among this set, we select a fraction of transcripts to be excluded from the annotation. These transcripts are denoted as the true novel isoforms.
To evaluate a transcript prediction tool, we provided the entire set of simulated reads and the reduced annotation as an input. Thus, true novel isoforms are hidden from the annotation, but present in the reads. We then compute precision and recall by running gffcomparefor the entire output annotation versus the complete set of expressed transcripts, reported known isoforms versus the set of true known isoforms and predicted novel transcript models versus the true novel set.
For the annotation-free benchmarks we simply compared the entire output annotation with the true set of expressed isoforms using gffcompare. To estimate how recall and precision of novel transcripts depend on the expression levels, predicted transcripts are grouped into bins by their transcripts per million values. For computing recall the number of false negative calls in each TPM bin is required. We thus group transcripts by their TPM values used during the simulation. However, computing precision requires the number of false-positive predictions within each bin and thus only reported TPM values can be used .
To evaluate SIRV transcripts we used an incomplete SIRV annotation containing only 43 out of 69 SIRV transcripts. The output annotations were again split into known and novel transcripts, and compared against the respective reference set using gffcompare. The SIRV-Set 4 annotations are available atConsistency between transcripts generated on real data was estimated using gffcompare .
France Dernières Nouvelles, France Actualités
Similar News:Vous pouvez également lire des articles d'actualité similaires à celui-ci que nous avons collectés auprès d'autres sources d'information.
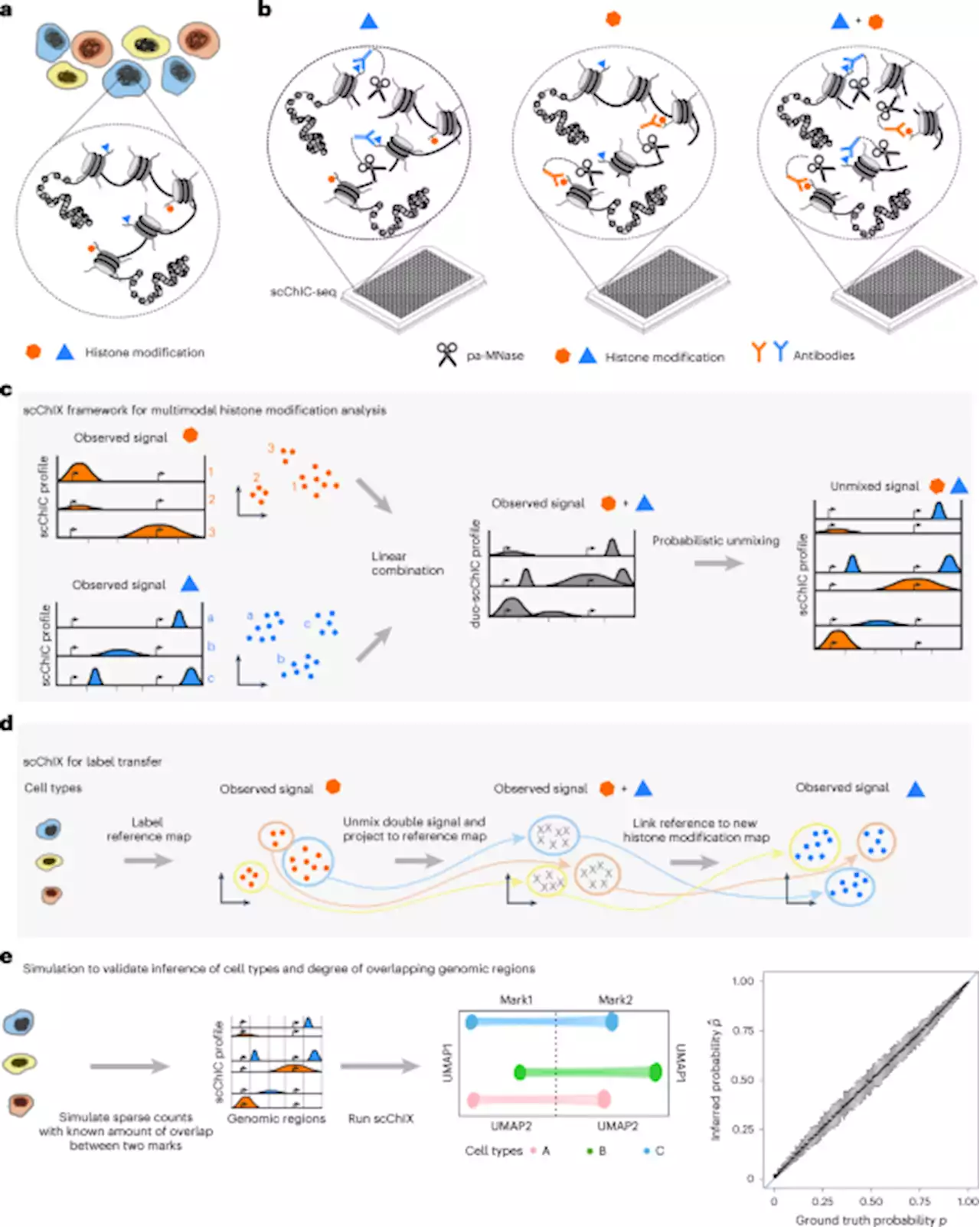 scChIX-seq infers dynamic relationships between histone modifications in single cells - Nature BiotechnologyAnalysis of two histone marks in single cells reveals interplay between modifications.
scChIX-seq infers dynamic relationships between histone modifications in single cells - Nature BiotechnologyAnalysis of two histone marks in single cells reveals interplay between modifications.
Lire la suite »
 T cell characteristics associated with toxicity to immune checkpoint blockade in patients with melanoma - Nature MedicineClonally diverse and activated memory CD4+ Tcells at baseline are associated with the development of severe irAEs in patients with melanoma treated with immune checkpoint inhibitors immunotherapy toxicity AaronNewmanLab aadel_chaudhuri ax_lozano
T cell characteristics associated with toxicity to immune checkpoint blockade in patients with melanoma - Nature MedicineClonally diverse and activated memory CD4+ Tcells at baseline are associated with the development of severe irAEs in patients with melanoma treated with immune checkpoint inhibitors immunotherapy toxicity AaronNewmanLab aadel_chaudhuri ax_lozano
Lire la suite »
 Ring Spotlight Cam Pro review | Digital TrendsThe Ring SpotlightCamPro uses radar for super-accurate motion detection, with advanced notification options that help cut down on notification spam.
Ring Spotlight Cam Pro review | Digital TrendsThe Ring SpotlightCamPro uses radar for super-accurate motion detection, with advanced notification options that help cut down on notification spam.
Lire la suite »
 Opinion | Making a case against Rep.-elect Santos could be harder than it seemsMatt Jacobs and Andrey Spektor: If Rep.-elect George Santos was as loose with disclosing his finances on campaign forms as he has been in public about his personal history, a federal indictment will be the likely result. - NBCNewsTHINK
Opinion | Making a case against Rep.-elect Santos could be harder than it seemsMatt Jacobs and Andrey Spektor: If Rep.-elect George Santos was as loose with disclosing his finances on campaign forms as he has been in public about his personal history, a federal indictment will be the likely result. - NBCNewsTHINK
Lire la suite »
 5 animal instincts that inspired technologyHere are five striking examples where nature has guided human innovation – and in some cases, could lead to even more exciting breakthroughs.
5 animal instincts that inspired technologyHere are five striking examples where nature has guided human innovation – and in some cases, could lead to even more exciting breakthroughs.
Lire la suite »